Building Custom CPOs
Martin Binder
2020-04-27
Source:vignettes/a_4_custom_CPOs.Rmd
a_4_custom_CPOs.RmdIntro
The CPOs built into mlrCPO can be used for many different purposes, and can be combined to form even more powerful transformation operations. However, in some cases, it may be necessary to define new “custom” CPOs that perform a certain task; either because a preprocessing method is not (yet) defined as a builtin CPO, or because some operation very specific to the task at hand needs to be performed.
For this purpose, mlrCPO offers a very powerful interface for the creation of new CPOs. The functions and methods described here are also the methods used internally to create mlrCPO’s builtin CPOs. Therefore, to learn the art of defining CPOs, it is also possible to look at the mlrCPO source tree in files starting with “CPO_” for example CPO definitions.
There are three types of CPO: “Feature Operation CPOs” (FOCPOs) which are only allowed to change feature columns of incoming data, and which are the most common CPOs; “Target Operation CPOs” (TOCPOs) that change only target columns, and “Retrafoless CPOs” (ROCPOs) that may add or delete rows to a data set, but only during training. Conceptually, ROCPOs are the simplest CPOs, followed by FOCPOs and the even more complicated TOCPOs. The commonalities of all CPO defining functions will be described first, followed by the different CPO types in order of growing complexity.
Making a CPO
To create a CPOConstructor that can then be used to create a CPO, a makeCPO*() function needs to be called. There are five functions of this kind, differing by what kind of CPO they create and how much flexibility (at the cost of simplicity) they offer the user:
CPO type |
makeCPO*() functions |
|---|---|
| FOCPO |
makeCPO(), makeCPOExtendedTrafo()
|
| TOCPO |
makeCPOTargetOp(), makeCPOExtendedTargetOp()
|
| ROCPO | makeCPORetrafoless() |
Each of these functions takes a “name” for the new CPO, settings for the parameter set to be used, settings for the format in which the data is supposed to be provided, data property settings, the packages to load, CPO type specific settins, and finally the transformation functions.
CPO name
Each CPO has a “name” that is used for representation when printing, and as the default prefix for hyperparameters. cpoPca, for example, has the name “pca”:
!cpoPca()
#> Trafo chain of 1 cpos:
#> pca(center = TRUE, scale = FALSE)[not exp'd: tol = <NULL>, rank = <NULL>]
#> Operating: feature
#> ParamSet:
#> Type len Def Constr Req Tunable Trafo
#> pca.center logical - TRUE - - TRUE -
#> pca.scale logical - FALSE - - TRUE -The name is set using the cpo.name parameter of the make*() functions.
CPO parameters
The ParSet used by the CPO are given as the second par.set parameter. These parameters must be either constructed using makeParamSet() from the ParamHelpers package, or using the pSS() function for a more concise ParSet definition. The given parameters will then be the function parameters of the CPOConstructor, and will by default be exported as hyperparameters (prefixed with the cpo.name).
It is possible to use the default parameter values of the par.set as defaults, or to give a par.vals list of default values. If par.vals is given, the defaults within par.set are completely ignored. Parameters that have a default value are set to this value upon construction if no value is given by the user.
Not all available parameters of a CPO need to be exported as hyperparameters. Which parameters are exported can be set during CPO construction, but the default exported parameters can be set using export.params. This can either be a character vector of the names of parameters to export, or TRUE (default, export all) or FALSE (no export).
Data Format
Different CPO operations may want to operate on the data in different forms: as a Task, as a data.frame with or without the target column, etc. The CPO framework can perform some conversion of data to fit different needs, which is set up by the value of fthe dataformat parameter, together with dataformat.factor.with.ordered. While dataformat has slightly different effects on different CPO types, typically its values and effects are:
dataformat |
Effect |
|---|---|
"task" |
Data is given as a Task; if the data to be transformed is a data.frame, it is converted to a cluster task before handing it to the transformation functions. |
"df.all" |
Data is given as a data.frame, with the target column included. |
"df.features" |
Data is given as a data.frame, the target is given as a separate data.frame. |
"split" |
Data is given as a named list with slots $numeric, $factor, $ordered, $other, each of which contains a data.frame with the columns of the respective type. If dataformat.factor.with.ordered is TRUE, the $ordered slot is not present, and ordered features are instead given to $factor as well. Features that are not any of these types are given to "other". The target is given as a separate data.frame. |
"factor", "ordered", "numeric"
|
Only the data from columns of the named type are given to the transformatin functions as a data.frame. The target columns are given as a separate data.frame. |
Another parameter influencing the data format is the fix.factors flag which controls whether factor levels of prediction data need to be set to be the same as during training. If it is TRUE, previously unseen factor levels are set to NA during prediction.
Properties
mlr and mlrCPO make it possible to specify what kind of data a CPO or a Learner can handle. However, since CPOs may change data to be more or less fitting for a certain Learner, a CPO must announce not only what data it can handle, but also how it changes the capabilities of the machine learning pipeline in which it is envolved. During construction, four parameters related to properties can be given.
The properties.data parameter defines what properties of feature data the CPO can handle; it must be a subset of "numerics", "factors", "ordered", and "missings". Typically, only the "missings" part is interesting since CPOs that only handle a subset of types will usually just ignore columns of other types.
The properties.target parameter defines what Task properties related to the task type and the target column a CPO can handle. It is a subset of "cluster", "classif", "multilabel", "regr", "surv" (so far defining the task type a CPO can handle), "oneclass", "twoclass", "multiclass" (properties specific to classif Tasks). Most FOCPOs do not care about the task type, while TOCPOs may only support a single task type.
properties.adding lists the properties that a CPO adds to the capabilities of a machine learning pipeline when it is executed before it, while properties.needed lists the properties needed from the following pipeline. cpoDummyEncode, for example, a CPO that converts factors and ordereds to numerics, has properties.adding == c("factors", "ordered") and properties.needed == "numerics". The many imputation CPOs have properties.adding == "missings". Usually these are only a subset of the possible properties.data states, but for TOCPOs this may also be any of "oneclass", "twoclass", "multiclass". Note that neither properties.adding nor properties.needed may be any task type, even for TOCPOs that perform task conversion.
Property Checking and .sometimes Properties
The CPO framework will check that a CPO only adds and removes the kind of data properties that it declared in properties.adding and properties.needed. It will also check that composition of CPOs, and attachment of CPOs to Learners, work out. Sometimes, however, it is necessary to treat a CPO like it does a certain manipulation (removing missings, for example) in some cases, while not in others. A CPO that only imputes missings in numeric columns should be treated as properties.adding == "missings" when is is attached to a Learner, and the Learner should gain the "missings" property. However, when data that has missings in its factorial columns is given to this CPO, the CPO framework will complain that the CPO that declared "missings" in properties.adding returned data that still had missing values in it. The solution to this dilemma is to suffix some properties with “.sometimes” when declaring them in properties.adding and properties.needed. When composing CPOs, and when checking data returned by a CPO, the framework will then be as lenient as possible. In the given example, properties.adding == "missings" will be assumed when attaching the CPO to a Learner, while properties.adding == character(0) is assumed when checking the CPO’s output (and missing values that were not imputed are therefore forgiven).
Packages
The single packages parameter can be set to a character vector listing packages necessary for a CPO to work. This is mostly useful when a CPO should be defined as part of a package or script to be distributed. The listed package will not automatically be attached, it will only be loaded. This means that a function exported by a package still needs to be called using ::. The benefit of declaring it in packages is that it will be loaded upon construction of a CPO, which means that a user will get immediate feedback about whether the CPO can be used or needs more packages to be installed.
Transformation Functions
The different types of CPO, and the different make*() functions, need different transformation functions to be defined. The principle behind these functions is alwasy the same, however: The CPO framework takes input data, transforms it according to dataformat, checks it according to properties.data and properties.target, and then gives it to one or more user-given transformation function. The transformation function must then usually create a control object containing information about the data to be used later, or transform the incoming data and return the transformation result (or both). The CPO framework then checks the transformed data according to properties.adding and properties.needed and gives it back to the CPO user.
Transformation functions are given to parameters starting with cpo.. They can either be given as functions, or as “headless” functions missing the function(...) part. In the latter case, the headless function must be a succession of expressions enclosed in curly braces ({, }) and the necessary function head is added by the CPO framework. The functions often take a subset of data, target, control, or control.invert parameters, in addition to all parameters as given in par.set.
Functional Transformation
The communication between transformation functions, e.g. giving the PCA matrix to its retrafo function, usually happens via “control” objects created by these functions and then given as parameter to other functions. In some cases, however, it may be more elegant to create a new function (e.g. a cpo.retrafo function) within another function as a “closure” (in the general, not R specific, sense) with access to all the outer functions variables. The CPO framework makes this possible by allowing a function to be given instead of a “control” object. The function which would usually receive this control object must then be given as NULL in the makeCPO*() call.
Retrafoless CPOs
Retrafoless CPOs, or ROCPOs, are conceptually the simplest CPO type, since they do not create CPOTrained objects and therefore only need one transformation function: cpo.trafo. The value of the dataformat parameter may only be either "df.all" or "task", resulting in either a data.frame (consisting all columns, including the target column) or a Task being given to the cpo.trafo function. cpo.trafo should have the parameters data (receiving the data as either a Task or data.frame), target (receiving the names of target columns in the data), and any parameter as given to par.set. The return value of cpo.trafo must be the transformed data, in the same format (data.frame or Task) as given as input.
Since a ROCPO only transforms incoming data during training, it should not do any transformation of target or feature values that would make it necessary to repeat this action during prediction. It may, for example, be used for subsampling a classification task to balance target classes, but it should not change the levels or values of given data rows.
The following is an example of a simplified version of the cpoSample CPO, which takes one parameter fraction and then subsamples a fraction part of incoming data without replacement:
xmpSample = makeCPORetrafoless("exsample", # nolint
pSS(fraction: numeric[0, 1]),
dataformat = "df.all",
cpo.trafo = function(data, target, fraction) {
newsize = round(nrow(data) * fraction)
row.indices = sample(nrow(data), newsize)
data[row.indices, ]
})
cpo = xmpSample(0.01)iris %>>% cpo
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 14 4.3 3.0 1.1 0.1 setosa
#> 50 5.0 3.3 1.4 0.2 setosaIt is possible to give the cpo.trafo as headless transformation function by just leaving out the function header. This can save a lot of boilerplate code when there are many parameters present, or when many transformation functions need to be given. The resulting CPO is completely equivalent to the one given above.
Feature Operation CPOs
FOCPOs are created with either the makeCPO() function, or the makeCPOExtendedTrafo() function. The former conceptually separates training from transformation, the latter separates transformation of training data from transformation of prediction data.
makeCPO()
In principle, a FOCPO needs a function that “trains” a control object depending on the data (cpo.train), and another function that uses this control object, and new data, to perform the preprocessing operation (cpo.retrafo). The cpo.train-function must return a “control” object which contains all information about how to transform a given dataset. cpo.retrafo takes a (potentially new!) dataset and the “control” object returned by cpo.trafo, and transforms the new data according to plan.
In contrast to makeCPORetrafoless(), the dataformat parameter of makeCPO() can take all values described in the section Data Format. The cpo.train function takes the arguments data, target, and any other parameter described in param.set. The data value is the incoming data as a Task, a data.frame with or without the target column, or a list of data.frames of different column types, according to dataformat. The target value is a character vector of target names if dataformat is "task" or "df.all", or a data.frame of the target columns otherwise.
The cpo.train function’s return value is treated as a control object and given to the cpo.retrafo function. Its parameters are data, control, and any parameters in par.set. The format of the data given to the data parameter is according to dataformat, with the exception that if dataformat is either "task" or "df.all", it will be treated here as if its value were "df.features". This is because the cpo.retrafo function is sometimes called with prediction data which does not have any target column at all.
It follows the simplified definition of a CPO that removes the numeric columns of smallest variance, returning a dataset of only n.col numeric columns. The dataformat variable is set to "numeric", so that only numeric columns are given to the CPO’s transformation functiosn; factorial columns are ignored. In cpo.trafo, calculates the variance of each of the data’s columns, and in cpo.retrafo it subsets the data according to these variances. Since cpo.retrafo may also be called during prediction with new data, the variance must not be calculated in cpo.retrafo–this could lead to cpo.retrafo filtering out different columns from cpo.trafo. This example also prints out which of its functions are being called.
xmpFilterVar = makeCPO("exemplvar", # nolint
pSS(n.col: integer[0, ]),
dataformat = "numeric",
cpo.train = function(data, target, n.col) {
cat("*** cpo.train ***\n")
sapply(data, var, na.rm = TRUE)
},
cpo.retrafo = function(data, control, n.col) {
cat("*** cpo.retrafo ***\n")
cat("Control:\n")
print(control)
cat("\n")
greatest = order(-control) # columns, ordered greatest to smallest var
data[greatest[seq_len(n.col)]]
})
cpo = xmpFilterVar(2)(Note that the function heads are optional.)
When the CPO is called with a dataset, the cpo.train function is called first, creating the control object which is then given to cpo.retrafo.
(trafd = head(iris) %>>% cpo)
#> *** cpo.train ***
#> *** cpo.retrafo ***
#> Control:
#> Sepal.Length Sepal.Width Petal.Length Petal.Width
#> 0.083000000 0.117666667 0.019000000 0.006666667
#> Species Sepal.Width Sepal.Length
#> 1 setosa 3.5 5.1
#> 2 setosa 3.0 4.9
#> 3 setosa 3.2 4.7
#> 4 setosa 3.1 4.6
#> 5 setosa 3.6 5.0
#> 6 setosa 3.9 5.4Note that the two columns of the entire iris dataset with the greatest variance are Petal.Length and Sepal.Length:
head(iris %>>% cpo)
#> *** cpo.train ***
#> *** cpo.retrafo ***
#> Control:
#> Sepal.Length Sepal.Width Petal.Length Petal.Width
#> 0.6856935 0.1899794 3.1162779 0.5810063
#> Species Petal.Length Sepal.Length
#> 1 setosa 1.4 5.1
#> 2 setosa 1.4 4.9
#> 3 setosa 1.3 4.7
#> 4 setosa 1.5 4.6
#> 5 setosa 1.4 5.0
#> 6 setosa 1.7 5.4However, when applying the retrafo() of trafd to the entire dataset, the same columns are filtered out as they were in the first transformation: Sepal.Width and Sepal.Length. When the retrafo() is used, cpo.train is not called; instead, the control object saved inside the retrafo is used.
head(iris %>>% retrafo(trafd))
#> *** cpo.retrafo ***
#> Control:
#> Sepal.Length Sepal.Width Petal.Length Petal.Width
#> 0.083000000 0.117666667 0.019000000 0.006666667
#> Species Sepal.Width Sepal.Length
#> 1 setosa 3.5 5.1
#> 2 setosa 3.0 4.9
#> 3 setosa 3.2 4.7
#> 4 setosa 3.1 4.6
#> 5 setosa 3.6 5.0
#> 6 setosa 3.9 5.4It is also possible to inspect the CPOTrained object to see that the control is there:
getCPOTrainedState(retrafo(trafd))
#> $n.col
#> [1] 2
#>
#> $control
#> Sepal.Length Sepal.Width Petal.Length Petal.Width
#> 0.083000000 0.117666667 0.019000000 0.006666667
#>
#> $data
#> $data$shapeinfo.input
#> <ShapeInfo (input) Sepal.Length: num, Sepal.Width: num, Petal.Length: num, Petal.Width: num, Species: fac>
#>
#> $data$shapeinfo.output
#> <ShapeInfo (output)>:
#> numeric:
#> <ShapeInfo Sepal.Width: num, Sepal.Length: num>
#> factor:
#> <ShapeInfo Species: fac>
#> other:
#> <ShapeInfo (empty)>Functional FOCPO
Instead of returning the control object, cpo.train may also return the cpo.retrafo function. This may be more succinct to write if there are many little pieces of information from the cpo.train run that the cpo.retrafo function should have access to.
When cpo.retrafo is given functionally, it should be a function with only one parameter: the newly incoming data. It can access the values of the par.set parameters from its encapsulating environment in cpo.train.
Note that the data and target values given to cpo.train are deleted after the cpo.train call, so cpo.retrafo does not have access to it. In fact, the CPO framework will give a warning about this.
xmpFilterVarFunc = makeCPO("exemplvar.func", # nolint
pSS(n.col: integer[0, ]),
dataformat = "numeric",
cpo.retrafo = NULL,
cpo.train = function(data, target, n.col) {
cat("*** cpo.train ***\n")
ctrl = sapply(data, var, na.rm = TRUE)
function(x) { # the data is given to the only present parameter: 'x'
cat("*** cpo.retrafo ***\n")
cat("Control:\n")
print(ctrl)
cat("\ndata:\n")
print(data) # 'data' is deleted: NULL
cat("target:\n")
print(target) # 'target' is deleted: NULL
greatest = order(-ctrl) # columns, ordered greatest to smallest var
x[greatest[seq_len(n.col)]]
}
})
cpo = xmpFilterVarFunc(2)(Note that the function heads are optional.)
(trafd = head(iris) %>>% cpo)
#> *** cpo.train ***
#> Warning in checkFunctionReturn(cpo.retrafo, "data", "cpo.retrafo", "cpo.train"): The function given as cpo.retrafo references a data and a target variable.
#> Beware that the 'data' and 'target' variable as given as an argument to the surrounding function
#> will not be accessible when cpo.retrafo is called.
#> If you still need to access this data, copy it to a variable with a different name.
#> If this warning is a false positive and you assign the 'data' variable properly, you can avoid
#> this warning by renaming the 'data' variable.
#> *** cpo.retrafo ***
#> Control:
#> Sepal.Length Sepal.Width Petal.Length Petal.Width
#> 0.083000000 0.117666667 0.019000000 0.006666667
#>
#> data:
#> NULL
#> target:
#> NULL
#> Species Sepal.Width Sepal.Length
#> 1 setosa 3.5 5.1
#> 2 setosa 3.0 4.9
#> 3 setosa 3.2 4.7
#> 4 setosa 3.1 4.6
#> 5 setosa 3.6 5.0
#> 6 setosa 3.9 5.4The CPOTrained state for a functional CPO is the environment of the retrafo function. It contains the “ctrl” variable defined during training, the parameters given to cpo.train, and the cpo.retrafo function itself. Note that data and target are deleted and replaced by different values.
getCPOTrainedState(retrafo(trafd))
#> $ctrl
#> Sepal.Length Sepal.Width Petal.Length Petal.Width
#> 0.083000000 0.117666667 0.019000000 0.006666667
#>
#> $data
#> $data$shapeinfo.input
#> <ShapeInfo (input) Sepal.Length: num, Sepal.Width: num, Petal.Length: num, Petal.Width: num, Species: fac>
#>
#> $data$shapeinfo.output
#> <ShapeInfo (output)>:
#> numeric:
#> <ShapeInfo Sepal.Width: num, Sepal.Length: num>
#> factor:
#> <ShapeInfo Species: fac>
#> other:
#> <ShapeInfo (empty)>
#>
#>
#> $target
#> NULL
#>
#> $n.col
#> [1] 2
#>
#> $cpo.retrafo
#> function(x) { # the data is given to the only present parameter: 'x'
#> cat("*** cpo.retrafo ***\n")
#> cat("Control:\n")
#> print(ctrl)
#> cat("\ndata:\n")
#> print(data) # 'data' is deleted: NULL
#> cat("target:\n")
#> print(target) # 'target' is deleted: NULL
#> greatest = order(-ctrl) # columns, ordered greatest to smallest var
#> x[greatest[seq_len(n.col)]]
#> }
#> <environment: 0xbf787c8>Stateless FOCPO
“Stateless” CPOs are CPOs that perform the same action during transformation of training and prediction data, independent from information during training. An example would be a CPO that converts all its columns to numeric columns. When a FOCPO does not need a state, the cpo.train parameter of makeCPO() can be set to NULL. The cpo.retrafo function then has no control paramter and instead only a data and any par.set parameter. The as.numeric-CPO could be written as the following:
xmpAsNum = makeCPO("asnum", # nolint
cpo.train = NULL,
cpo.retrafo = function(data) {
data.frame(lapply(data, as.numeric))
})
cpo = xmpAsNum()(Note that the function head is optional.)
(trafd = head(iris) %>>% cpo)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 5.1 3.5 1.4 0.2 1
#> 2 4.9 3.0 1.4 0.2 1
#> 3 4.7 3.2 1.3 0.2 1
#> 4 4.6 3.1 1.5 0.2 1
#> 5 5.0 3.6 1.4 0.2 1
#> 6 5.4 3.9 1.7 0.4 1The “state” of the CPOTrained object thus created only contains information about the incoming data shape, to make sure that the CPOTrained object is only used on conforming data (as doing otherwise would indicate a bug).
getCPOTrainedState(retrafo(trafd))
#> $data
#> $data$shapeinfo.input
#> <ShapeInfo (input) Sepal.Length: num, Sepal.Width: num, Petal.Length: num, Petal.Width: num, Species: fac>
#>
#> $data$shapeinfo.output
#> <ShapeInfo (output) Sepal.Length: num, Sepal.Width: num, Petal.Length: num, Petal.Width: num, Species: num>
makeCPOExtendedTrafo()
Sometimes it is advantageous to have the training operation return the transformed data right away. PCA, for example, returns the rotation matrix and the transformed data; it would be a waste of time to only return the rotation matrix in a cpo.train function and apply it on the training data in cpo.retrafo. The makeCPOExtendedTrafo() function works very much like makeCPO(), with the difference that it has a cpo.trafo instead of a cpo.train function parameter. The cpo.trafo takes the same parameters as cpo.train, but returns the transformed data instead of a control object. The control object needs to be created additionally, as a variable by the cpo.trafo function. The CPO framework takes the value of a variable named control inside the cpo.trafo function and gives it to the cpo.retrafo function.
The following is a simplified version of the cpoPca CPO, which does not scale or center the data.
xmpPca = makeCPOExtendedTrafo("simple.pca", # nolint
pSS(n.col: integer[0, ]),
dataformat = "numeric",
cpo.trafo = function(data, target, n.col) {
cat("*** cpo.trafo ***\n")
pcr = prcomp(as.matrix(data), center = FALSE, scale. = FALSE, rank = n.col)
# save the rotation matrix as 'control' variable
control = pcr$rotation
pcr$x
},
cpo.retrafo = function(data, control, n.col) {
cat("*** cpo.retrafo ***\n")
# rotate the data by the rotation matrix
as.matrix(data) %*% control
})
cpo = xmpPca(2)When this CPO is applied to data, only the cpo.trafo function is called.
(trafd = head(iris) %>>% cpo)
#> *** cpo.trafo ***
#> Species PC1 PC2
#> 1 setosa -6.344251 3.699099e-05
#> 2 setosa -5.909522 -2.939100e-01
#> 3 setosa -5.835572 -1.780612e-02
#> 4 setosa -5.747518 -5.192580e-02
#> 5 setosa -6.319018 1.359890e-01
#> 6 setosa -6.882318 1.859359e-01When the retrafo CPOTrained is used, the cpo.retrafo function is called, making use of the rotation matrix.
tail(iris) %>>% retrafo(trafd)
#> *** cpo.retrafo ***
#> Species PC1 PC2
#> 145 virginica -8.614333 -0.7582980
#> 146 virginica -8.324575 -1.0203095
#> 147 virginica -7.667317 -1.2766319
#> 148 virginica -8.152840 -0.9708329
#> 149 virginica -8.190484 -0.4297855
#> 150 virginica -7.640937 -0.6764400The rotation matrix can be inspected using getCPOTrainedState.
getCPOTrainedState(retrafo(trafd))
#> $n.col
#> [1] 2
#>
#> $control
#> PC1 PC2
#> Sepal.Length -0.8012756 -0.55116118
#> Sepal.Width -0.5489467 0.80835852
#> Petal.Length -0.2348418 -0.04199971
#> Petal.Width -0.0382656 0.20251883
#>
#> $data
#> $data$shapeinfo.input
#> <ShapeInfo (input) Sepal.Length: num, Sepal.Width: num, Petal.Length: num, Petal.Width: num, Species: fac>
#>
#> $data$shapeinfo.output
#> <ShapeInfo (output)>:
#> numeric:
#> <ShapeInfo PC1: num, PC2: num>
#> factor:
#> <ShapeInfo Species: fac>
#> other:
#> <ShapeInfo (empty)>Functional FOCPO
As with makeCPO(), makeCPOExtendedTrafo() makes it possible to define functional CPOs. Instead of returning a cpo.retrafo function, the cpo.retrafo function needs to be defined as a variable, instead of a “control” variable. Like in makeCPO(), the cpo.retrafo parameter of makeCPOExtendedTrafo() must then be NULL. The PCA example above could thus also be written as
xmpPcaFunc = makeCPOExtendedTrafo("simple.pca.func", # nolint
pSS(n.col: integer[0, ]),
dataformat = "numeric",
cpo.retrafo = NULL,
cpo.trafo = function(data, target, n.col) {
cat("*** cpo.trafo ***\n")
pcr = prcomp(as.matrix(data), center = FALSE, scale. = FALSE, rank = n.col)
# save the rotation matrix as 'control' variable
cpo.retrafo = function(data) {
cat("*** cpo.retrafo ***\n")
# rotate the data by the rotation matrix
as.matrix(data) %*% pcr$rotation
}
pcr$x
})
cpo = xmpPcaFunc(2)(trafd = head(iris) %>>% cpo)
#> *** cpo.trafo ***
#> Species PC1 PC2
#> 1 setosa -6.344251 3.699099e-05
#> 2 setosa -5.909522 -2.939100e-01
#> 3 setosa -5.835572 -1.780612e-02
#> 4 setosa -5.747518 -5.192580e-02
#> 5 setosa -6.319018 1.359890e-01
#> 6 setosa -6.882318 1.859359e-01This also serves as an example of the disadvantages of a functional CPO: Since the CPO state contains all the information contained in the cpo.trafo call (except the data and target variables), it may take up more memory than needed. For this CPO, the state contains the pcr variable which contains the transformed training data in its $x slot. If the training data is a very large dataset, this would result in CPO states that take up a lot of working memory.
Target Operation CPOs
TOCPOs are more complicated than FOCPOs, since they potentially need to operate on data at three different points: During initial training, during the re-transformation for new prediction data, and during the inversion of predictions made by a model trained on transformed data. Similarly to makeCPO(), makeCPOTargetOp() splits these operations up into functions that create “control” objects, and functions that do the actual transformation. makeCPOExtendedTargetOp(), on the other hand, gives the user more flexibility at the price of the user having to make sure that transformation and retransformation perform the same operation–similarly to makeCPOExtendedTrafo() for FOCPOs.
Task Type and Conversion
In contrast to FOCPOs, TOCPOs can only operate on one type of Task. Therefore, the properties.target parameter of makeCPO*TargetOp() must contain exactly one Task type ("cluster", "classif", "regr", "surv", "multilabel") and possibly some more task properties (currently only "oneclass", "twoclass", "multiclass" if the Task type is "classif").
It is possible to write TOCPOs that perform conversion of Task types. For that, the task.type.out parameter must be set to the Task type that the CPO converts the data to. If conversion happens, the transformation functions need to return target data fit for the task.type.out Task type.
properties.adding and properties.needed should not be any Task type, even when conversion happens. Only if one of the task types has additional properties–currently only the "oneclass", "twoclass", "multiclass" properties of classification Tasks–should these additional properties be listed in properties.adding or properties.needed.
predict.type
mlr makes it possible for Learners to make different kinds of prediction. Usually they can predict a “response”, making their best effort to predict the true value of a task target. Many Learner types can predict a probability when their predict.type is set to "prob", returning a data.frame of their estimated probability distribution over possible responses. For regression Learners, predict.type can be "se" for the Learner to predict its estimated standard error of their response prediction.
When TOCPOs invert these predictions, they may
- declare which kind of
predict.typepredictions they can perform - declare what
predict.typethey require from the underlyingLearnerto make thispredict.typeprediction.
This is done using the predict.type.map parameter of makeCPO*TargetOp(). It is a named list or named character vector with the names indicating the supported predict.types, and the values indicating the required underlying predictions. For example, if a TOCPO can perform "response" and "se" prediction, and to predict "response" the underlying Learner must also perform "response" prediction, but for "se" prediction it must perform "prob" prediction, the predict.type.map would have the value
makeCPOTargetOp()
makeCPOTargetOp() has a cpo.train and cpo.retrafo function parameter that work similarly to the ones of makeCPO(). In contrast to makeCPO(), however, cpo.retrafo must return the target data instead of the feature data. The data and target parameters of cpo.retrafo get the same data as they get in a FOCPO created with makeCPO(), with the exception that if dataformat is "task" or "df.all", the target parameter will receive the whole input data in form of a Task or data.frame (while the data argument, as in a FOCPO, will receive only the feature data.frame). The return value of cpo.retrafo for a TOCPO must always be in the same format as the input target value: a data.frame with the manipulated target values when dataformat is anything besides "task" or "df.all", or a Task or data.frame of all data (with non-target columns unmodified) otherwise.
Inversion of predictions is performed using the functions cpo.train.invert and cpo.invert. cpo.train.invert takes a data and a control argument, and any arguments declared in the par.set. It is called whenever new data is fed into the CPO or its retrafo CPOTrained, and creates a CPOTrained state that is used to invert the prediction done on this new data. The control argument takes the value returned by the cpo.train function upon initial training, and the data argument is the new data for which to prepare the CPOTrained inverter. It has the form dictated by dataformat, with the exception that "task" and "df.all" dataformat are handled as "df.feature"; this is necessary since the new data could be a data.frame of data with unknown target.
The following is an example of a TOCPO that trains a classification Learner on a binary classification Task and changes it to a Task of whether or not the Learner predicted the truth for a given data line correctly. (Real-world applications would probably need to take some precautions against overfitting.) In its cpo.train step, the given Learner is trained on the incoming data and the resulting WrappedModel object is returned as the “control” object. This is given to the cpo.retrafo function, which performs prediction and creates a new classification Task with the match / mismatch between model prediction and ground truth as target. When an external Learner is trained on data that was preprocessed like this, its prediction will be whether the CPO-internal Learner can be trusted to predict a given data row. To “invert” this, i.e. to get the actual prediction, the cpo.invert function needs to have the internal Learner’s prediction as well as the prediction made by the external Learner. The former is provided by cpo.train.invert, which uses the WrappedModel to make a prediction on the new data, and given as control.invert to cpo.invert. The latter is the target data given to cpo.invert. This example CPO supports inverting both "response" and "prob" predict.type predictions, as declared in the predict.type.map argument. The actual predict.type to invert is given to cpo.invert as an argument.
xmpMetaLearn = makeCPOTargetOp("xmp.meta", # nolint
pSS(lrn: untyped),
dataformat = "task",
properties.target = c("classif", "twoclass"),
predict.type.map = c(response = "response", prob = "prob"),
cpo.train = function(data, target, lrn) {
cat("*** cpo.train ***\n")
lrn = setPredictType(lrn, "prob")
train(lrn, data)
},
cpo.retrafo = function(data, target, control, lrn) {
cat("*** cpo.retrafo ***\n")
prediction = predict(control, target)
tname = getTaskTargetNames(target)
tdata = getTaskData(target)
tdata[[tname]] = factor(prediction$data$response == prediction$data$truth)
makeClassifTask(getTaskId(target), tdata, tname, positive = "TRUE",
fixup.data = "no", check.data = FALSE)
},
cpo.train.invert = function(data, control, lrn) {
cat("*** cpo.train.invert ***\n")
predict(control, newdata = data)$data
},
cpo.invert = function(target, control.invert, predict.type, lrn) {
cat("*** cpo.invert ***\n")
if (predict.type == "prob") {
outmat = as.matrix(control.invert[grep("^prob\\.", names(control.invert))])
revmat = outmat[, c(2, 1)]
outmat * target[, "prob.TRUE", drop = TRUE] +
revmat * target[, "prob.FALSE", drop = TRUE]
} else {
stopifnot(levels(target) == c("FALSE", "TRUE"))
numeric.prediction = as.numeric(control.invert$response)
numeric.res = ifelse(target == "TRUE",
numeric.prediction,
3 - numeric.prediction)
factor(levels(control.invert$response)[numeric.res],
levels(control.invert$response))
}
})
cpo = xmpMetaLearn(makeLearner("classif.logreg"))To show the inner workings of this CPO, the following example data is used.
set.seed(12)
split = makeResampleInstance(hout, pid.task)
train.task = subsetTask(pid.task, split$train.inds[[1]])
test.task = subsetTask(pid.task, split$predict.inds[[1]])It can be instructive to watch the cat() output of this CPO to see which function gets called at what point in the lifecycle. The cpo.train function is called first to create the control object. The Task is transformed in cpo.retrafo. Also cpo.train.invert is called, since an inverter attribute is attached to the returned trafo.
trafd = train.task %>>% cpo
#> *** cpo.train ***
#> *** cpo.train.invert ***
#> *** cpo.retrafo ***
attributes(trafd)
#> $names
#> [1] "type" "env" "weights" "blocking" "coordinates"
#> [6] "task.desc"
#>
#> $class
#> [1] "ClassifTask" "SupervisedTask" "Task"
#>
#> $retrafo
#> CPO Retrafo chain {type:classif}
#> [RETRAFO xmp.meta(lrn = <classif.log...)]
#>
#> $inverter
#> CPO Inverter chain {type:classif} (able to predict 'response', 'prob')
#> [INVERTER xmp.meta(lrn = <classif.log...){type:classif}]The values of the target column (“diabetes”) of the result can be compared with the prediction of a "classif.logreg" Learner on the same data:
head(getTaskData(trafd))
#> pregnant glucose pressure triceps insulin mass pedigree age diabetes
#> 450 0 120 74 18 63 30.5 0.285 26 TRUE
#> 346 8 126 88 36 108 38.5 0.349 49 FALSE
#> 336 0 165 76 43 255 47.9 0.259 26 FALSE
#> 247 10 122 68 0 0 31.2 0.258 41 TRUE
#> 174 1 79 60 42 48 43.5 0.678 23 TRUE
#> 453 0 91 68 32 210 39.9 0.381 25 TRUEmodel = train(makeLearner("classif.logreg", predict.type = "prob"), train.task)
head(predict(model, train.task)$data[c("truth", "response")])
#> truth response
#> 450 neg neg
#> 346 neg pos
#> 336 neg pos
#> 247 neg neg
#> 174 neg neg
#> 453 neg negWhen new data is transformed using the retrafo CPOTrained, another inverter attribute is created, and hence cpo.train.invert is called again. Since the target column of the test.task in the following example is also transformed, the cpo.retrafo function is called.
retr = test.task %>>% retrafo(trafd)
#> *** cpo.retrafo ***
#> *** cpo.train.invert ***
attributes(retr)
#> $names
#> [1] "type" "env" "weights" "blocking" "coordinates"
#> [6] "task.desc"
#>
#> $class
#> [1] "ClassifTask" "SupervisedTask" "Task"
#>
#> $inverter
#> CPO Inverter chain {type:classif} (able to predict 'response', 'prob')
#> [INVERTER xmp.meta(lrn = <classif.log...){type:classif}]In a real world application, it would be possible for the new incoming data to have unknown target values. In that case, no target column would need to be changed, and cpo.retrafo is not called. The resulting data, retr.df, equals the input data with a retrafo attribute added.
retr.df = getTaskData(test.task, target.extra = TRUE)$data %>>% retrafo(trafd)
#> *** cpo.train.invert ***
names(attributes(retr.df))
#> [1] "names" "class" "row.names" "inverter"The invert functionality can be demonstrated by making a prediction with an external model.
ext.model = train("classif.svm", trafd)
ext.pred = predict(ext.model, retr)
newpred = invert(inverter(retr), ext.pred)
#> *** cpo.invert ***
performance(newpred)
#> mmce
#> 0.2200521It may also be instructive to attach the xmpMetaLearn CPO to a Learner to see which functions get called during training and prediction of a TOCPO-Learner. Since the Learner does not do inversion of the training data, a CPOTrained for inversion is not created during training, and cpo.train.invert is hence not called. Only cpo.train (for control object creation) and cpo.retrafo (target value change) are called. During prediction, the input data is used to create an (internally used) inversion CPOTrained which promptly gets used by the prediction made by "classif.svm". Hence both cpo.train.invert and cpo.invert are called in succession.
cpo.learner = cpo %>>% makeLearner("classif.svm")
cpo.model = train(cpo.learner, train.task)
#> *** cpo.train ***
#> *** cpo.retrafo ***lrnpred = predict(cpo.model, test.task)
#> *** cpo.train.invert ***
#> *** cpo.invert ***
performance(lrnpred)
#> mmce
#> 0.2200521See Postscriptum for an evaluation of xmpMeatLearn’s performance.
Functional TOCPO
Just like for FOCPOs, it is possible to create functional TOCPOs. In the case of makeCPOTargetOp(), it is possible to have cpo.train create cpo.retrafo and cpo.train.invert, instead of giving them to makeCPOTargetOp() directly. Just as in makeCPO, these functions can then access the state of their environment in the cpo.train call and hence have neither a control argument, nor any arguments for the par.set parameters. Since cpo.train must in this case create two functions, these functions only need to be defined within cpo.train, the return value is ignored.
Note that cpo.retrafo and cpo.train.invert must either be both functional or both object based.
It is furthermore possible to return a cpo.invert function by cpo.train.invert, instead of giving it to makeCPOTargetOp(). As above, the returned function should not have any parameters for the ones given in par.set, and should not have a control.invert. cpo.invert can be functional or not, independently of whether cpo.retrafo and cpo.train.invert are functional.
As in makeCPO(), all functions that are given functionally must be explicitly set to NULL in the makeCPOTargetOp() call.
The xmpMetaLearn example above with functional cpo.retrafo, cpo.train.invert and cpo.invert would look like the following:
xmpMetaLearn = makeCPOTargetOp("xmp.meta.fnc", # nolint
pSS(lrn: untyped),
dataformat = "task",
properties.target = c("classif", "twoclass"),
predict.type.map = c(response = "response", prob = "prob"),
# set the cpo.* parameters not needed to NULL:
cpo.retrafo = NULL, cpo.train.invert = NULL, cpo.invert = NULL,
cpo.train = function(data, target, lrn) {
cat("*** cpo.train ***\n")
lrn = setPredictType(lrn, "prob")
model = train(lrn, data)
cpo.retrafo = function(data, target) {
cat("*** cpo.retrafo ***\n")
prediction = predict(model, target)
tname = getTaskTargetNames(target)
tdata = getTaskData(target)
tdata[[tname]] = factor(prediction$data$response == prediction$data$truth)
makeClassifTask(getTaskId(target), tdata, tname, positive = "TRUE",
fixup.data = "no", check.data = FALSE)
}
cpo.train.invert = function(data) {
cat("*** cpo.train.invert ***\n")
prediction = predict(model, newdata = data)$data
function(target, predict.type) { # this is returned as cpo.invert
cat("*** cpo.invert ***\n")
if (predict.type == "prob") {
outmat = as.matrix(prediction[grep("^prob\\.", names(prediction))])
revmat = outmat[, c(2, 1)]
outmat * target[, "prob.TRUE", drop = TRUE] +
revmat * target[, "prob.FALSE", drop = TRUE]
} else {
stopifnot(levels(target) == c("FALSE", "TRUE"))
numeric.prediction = as.numeric(prediction$response)
numeric.res = ifelse(target == "TRUE",
numeric.prediction,
3 - numeric.prediction)
factor(levels(prediction$response)[numeric.res],
levels(prediction$response))
}
}
}
})Constant Invert TOCPOs
The example given above is a relatively elaborate TOCPO which needs information from the prediction data to perform inversion. Many simpler applications of target transformation do not need this information if their inversion step is independent of this data. It is possible to declare such a TOCPO using the constant.invert flag in makeCPOTargetOp(). If constant.invert is set to TRUE, the cpo.train.invert argument must be explicitly set to NULL. cpo.train still needs to have a control.invert argument; it is set to the value returned by cpo.train.
The following example is a TOCPO for regression Tasks that centers target values during training. After prediction, the data is inverted by adding the original mean of the training data to the predictions. This inversion operation does not need any information about the prediction data going in, so the TOCPO can be declared constant.invert.
The cpo.retrafo function is also called when new prediction data with a target column is transformed (as during model validation). In that case, the mean of the training data column is subtracted. Therefore the mean generated by cpo.train needs to be used in cpo.retrafo (i.e. the control value), not the mean of the target data present.
xmpRegCenter = makeCPOTargetOp("xmp.center", # nolint
constant.invert = TRUE,
cpo.train.invert = NULL, # necessary for constant.invert = TRUE
dataformat = "df.feature",
properties.target = "regr",
cpo.train = function(data, target) {
# control value is just the mean of the target column
mean(target[[1]])
},
cpo.retrafo = function(data, target, control) {
# subtract mean from target column in retrafo
target[[1]] = target[[1]] - control
target
},
cpo.invert = function(target, predict.type, control.invert) {
target + control.invert
})
cpo = xmpRegCenter()To illustrate this CPO, the following data is used:
train.task = subsetTask(bh.task, 150:155)
getTaskTargets(train.task)
#> [1] 15.4 21.5 19.6 15.3 19.4 17.0predict.task = subsetTask(bh.task, 156:160)
getTaskTargets(predict.task)
#> [1] 15.6 13.1 41.3 24.3 23.3The target column of the task after transformation has a mean of 0.
trafd = train.task %>>% cpo
getTaskTargets(trafd)
#> [1] -2.633333 3.466667 1.566667 -2.733333 1.366667 -1.033333When applying the retrafo CPOTrained to a new task, the mean of the training task target column is subtracted.
retr = retrafo(trafd)
predict.traf = predict.task %>>% retr
getTaskTargets(predict.traf)
#> [1] -2.433333 -4.933333 23.266667 6.266667 5.266667When inverting a regression prediction, the mean of the training data target column is added to the prediction.
model = train("regr.lm", trafd)
pred = predict(model, predict.traf)
#> Warning in predict.lm(.model$learner.model, newdata = .newdata, se.fit =
#> FALSE, : prediction from a rank-deficient fit may be misleading
pred
#> Prediction: 5 observations
#> predict.type: response
#> threshold:
#> time: 0.00
#> id truth response
#> 156 1 -2.433333 -2.854142
#> 157 2 -4.933333 5.802064
#> 158 3 23.266667 14.442103
#> 159 4 6.266667 8.302782
#> 160 5 5.266667 8.633308invert(inverter(predict.traf), pred)
#> Prediction: 5 observations
#> predict.type: response
#> threshold:
#> time: 0.00
#> id truth response
#> 156 1 15.6 15.17919
#> 157 2 13.1 23.83540
#> 158 3 41.3 32.47544
#> 159 4 24.3 26.33611
#> 160 5 23.3 26.66664Since "regr.lm" is translation invariant and deterministic, the prediction equals the prediction made without centering the target:
model = train("regr.lm", train.task)
predict(model, predict.task)
#> Warning in predict.lm(.model$learner.model, newdata = .newdata, se.fit =
#> FALSE, : prediction from a rank-deficient fit may be misleading
#> Prediction: 5 observations
#> predict.type: response
#> threshold:
#> time: 0.00
#> id truth response
#> 156 1 15.6 15.17919
#> 157 2 13.1 23.83540
#> 158 3 41.3 32.47544
#> 159 4 24.3 26.33611
#> 160 5 23.3 26.66664A special property of constant.invert TOCPOs is that their retrafo CPOTrained can also be used for inversion. This is the case since the tight coupling of inversion operation to the data used to create the prediction is not necessary when the inversion is actually independent of this data. This is indicated by getCPOTrainedCapability() returning a vector with the "invert" capability set to 1. However, when using the retrafo CPOTrained for inversion, the “truth” column is absent from the inverted prediction.
Functional Constant Invert TOCPO
Just as above, constant.invert TOCPOs can be functional. For this, the cpo.train function must declare both a cpo.retrafo and a cpo.invert variable which perform the requested operations. These functions have no control or control.invert parameter, and no parameters pertaining to par.set.
Stateless TOCPO
Very simple target column operations that operate on a row-by-row basis without needing information e.g. from training data, can be declared as “stateless”. Similarly to makeCPO(), when cpo.train parameter is set to NULL, no control object is created for a CPOTrained. Furthermore, a stateless TOCPO must always have constant.invert set as well. Therefore, only cpo.retrafo and cpo.invert are given as functions, both without a control or control.invert argument. One example is a TOCPO that log-transforms the target column of a regression task, and exponentiates the predictions made from this during inversion. (A better inversion would take the "se" prediction into account, see cpoLogTrafoRegr.)
xmpLogRegr = makeCPOTargetOp("log.regr", # nolint
constant.invert = TRUE,
properties.target = "regr",
cpo.train = NULL, cpo.train.invert = NULL,
cpo.retrafo = function(data, target) {
target[[1]] = log(target[[1]])
target
},
cpo.invert = function(target, predict.type) {
exp(target)
})
cpo = xmpLogRegr()The CPO takes the logarithm of the task target column both during training and when using the retrafo CPOTrained.
trafd = train.task %>>% cpo
getTaskTargets(trafd)
#> [1] 2.734368 3.068053 2.975530 2.727853 2.965273 2.833213retr = retrafo(trafd)
predict.traf = predict.task %>>% retr
getTaskTargets(predict.traf)
#> [1] 2.747271 2.572612 3.720862 3.190476 3.148453model = train("regr.lm", trafd)
pred = predict(model, predict.traf)
#> Warning in predict.lm(.model$learner.model, newdata = .newdata, se.fit =
#> FALSE, : prediction from a rank-deficient fit may be misleading
pred
#> Prediction: 5 observations
#> predict.type: response
#> threshold:
#> time: 0.00
#> id truth response
#> 156 1 2.747271 2.726065
#> 157 2 2.572612 3.224557
#> 158 3 3.720862 3.685047
#> 159 4 3.190476 3.353014
#> 160 5 3.148453 3.365374Note that both the inverter and the retrafo CPOTrained can be used for inversion, since a stateless TOCPO also has constant.invert set. As above, when using the retrafo CPOTrained, the truth column is absent from the result.
makeCPOExtendedTargetOp()
Just as for FOCPOs, it is possible to declare a TOCPO while having more direct control over what happens at which stage of training, re-transformation, or inversion. In a TOCPO defined with makeCPOTargetOp(), the cpo.retrafo and cpo.train.invert functions are called automatically when necessary during training and re-transformation. makeCPOExtendedTargetOp() instead has a cpo.trafo and a cpo.retrafo parameter, which get called during the respective operation.
cpo.trafo must be a function taking the same parameters as cpo.train in makeCPOTargetOp(). Instead of returning a control object, it must define a variable named “control”, and a variable named “control.invert”. The former is used as the control argument of cpo.retrafo, the latter is used as control.invert for cpo.invert when using the inverter CPOTrained created during training. The return value of cpo.trafo must be similar to the value returned by cpo.retrafo in makeCPOTargetOp(): it must be the modified data set or target, depending on dataformat.
cpo.retrafo must take the same parameters as in makeCPOTargetOp(). It must declare a control.invert variable that will be given to cpo.retrafo when using the inverter CPOTrained created during retransformation. Since cpo.retrafo is always called during retrafo CPOTrained application, a “target” column may or may not be present. If a target column is not present, the target parameter of cpo.retrafo is NULL and the return value of cpo.retrafo is ignored; otherwise it must be the transformed target value (which, as in makeCPOTargetOp(), can be a Task or data.frame of all data if dataformat is "task" or "df.all").
cpo.invert works just as in makeCPOTargetOp().
The following is a nonsensical, synthetic example that adds 1 to the target column of a regression Task during initial training, subtracts 1 during retrafo re-application and is a no-op during inversion.
xmpSynCPO = makeCPOExtendedTargetOp("syn.cpo", # nolint
properties.target = "regr",
cpo.trafo = function(data, target) {
cat("*** cpo.trafo ***\n")
target[[1]] = target[[1]] + 1
control = "control created in cpo.trafo"
control.invert = "control.invert created in cpo.trafo"
target
},
cpo.retrafo = function(data, target, control) {
cat("*** cpo.retrafo ***", "control is:", deparse(control), sep = "\n")
control.invert = "control.invert created in cpo.retrafo"
if (!is.null(target)) {
cat("target is non-NULL, performing transformation\n")
target[[1]] = target[[1]] - 1
return(target)
} else {
cat("target is NULL, no transformation (but control.invert was created)\n")
return(NULL) # is ignored.
}
},
cpo.invert = function(target, control.invert, predict.type) {
cat("*** invert ***", "control.invert is:", deparse(control.invert),
sep = "\n")
target
})
cpo = xmpSynCPO()For an “extended” TOCPO, only one of the transformation functions is called in each invocation. Initial transformation calls cpo.trafo and adds 1 to the targets; using the CPOTrained for re-transformation calls cpo.retrafo and subtracts 1.
trafd = train.task %>>% cpo
#> *** cpo.trafo ***
getTaskTargets(trafd)
#> [1] 16.4 22.5 20.6 16.3 20.4 18.0retrafd = train.task %>>% retrafo(trafd)
#> *** cpo.retrafo ***
#> control is:
#> "control created in cpo.trafo"
#> target is non-NULL, performing transformationIt is also possible to perform re-transformation with a data.frame that does not include the target column. In that case the target value given to cpo.retrafo will be NULL, as reported by that function in this example:
retrafd = getTaskData(train.task, target.extra = TRUE)$data %>>% retrafo(trafd)
#> *** cpo.retrafo ***
#> control is:
#> "control created in cpo.trafo"
#> target is NULL, no transformation (but control.invert was created)The trafd object has an inverter CPOTrained attribute that was created by cpo.trafo, the retrafd object has an inverter CPOTrained attribute created by cpo.retrafo (necessarily). This is made visible by the given example inverter function:
Postscriptum
As an aside, the Learner enhanced by xmpMetaLearn seems to perform marginally better than either "classif.svm" or "classif.logreg" on their own for a large enough subset of pid.task (here resampled with output suppressed).
learners = list(
logreg = makeLearner("classif.logreg"),
svm = makeLearner("classif.svm"),
cpo = xmpMetaLearn(makeLearner("classif.logreg")) %>>%
makeLearner("classif.svm")
)
# suppress output of '*** cpo.train ***' etc.
configureMlr(show.info = FALSE, show.learner.output = FALSE)
perfs = sapply(learners, function(lrn) {
unname(replicate(20, resample(lrn, pid.task, cv10)$aggr))
})
# reset mlr settings
configureMlr()
boxplot(perfs)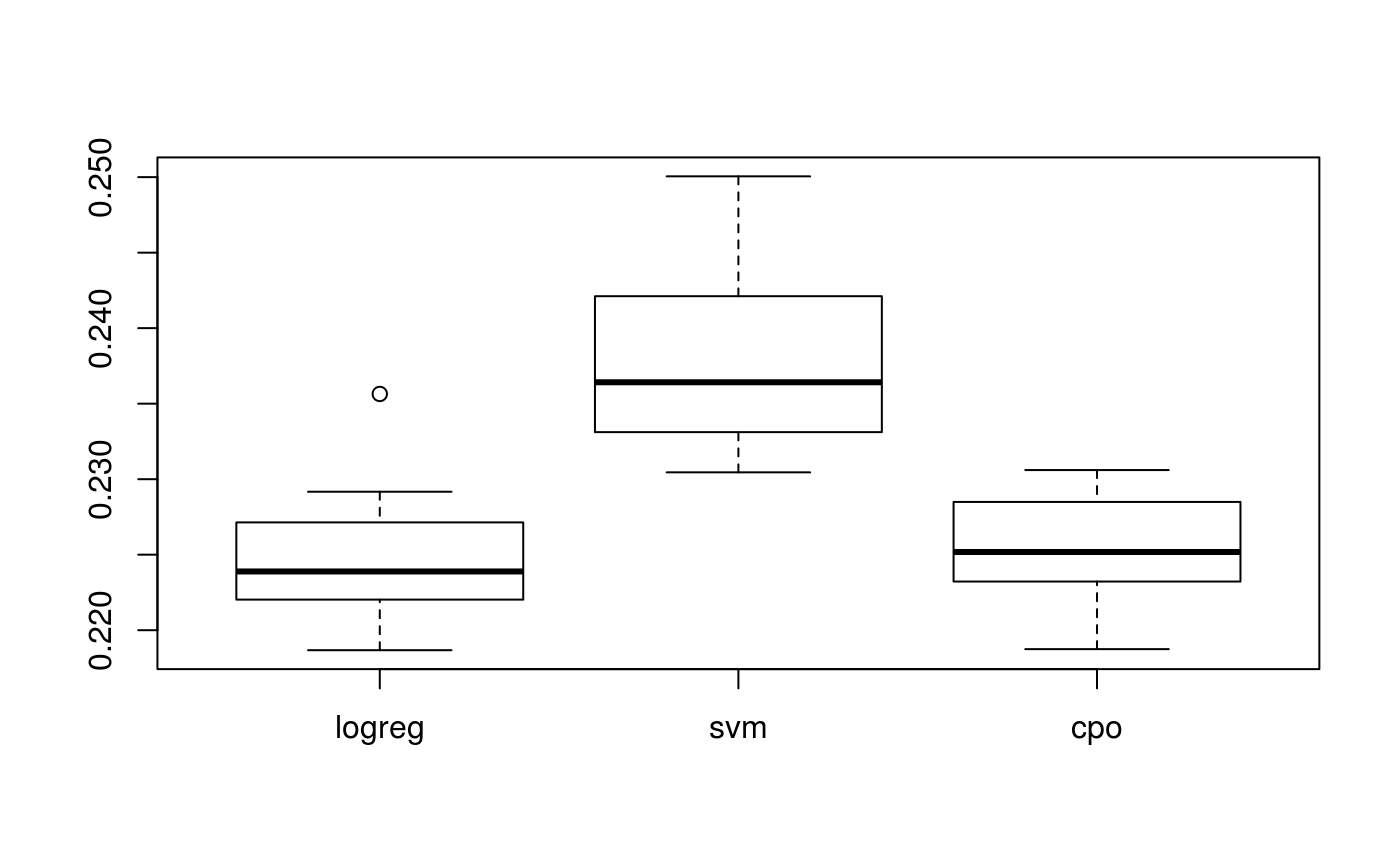
P-Values of comparing the CPOLearner to both "classif.logreg", and "classif.svm":